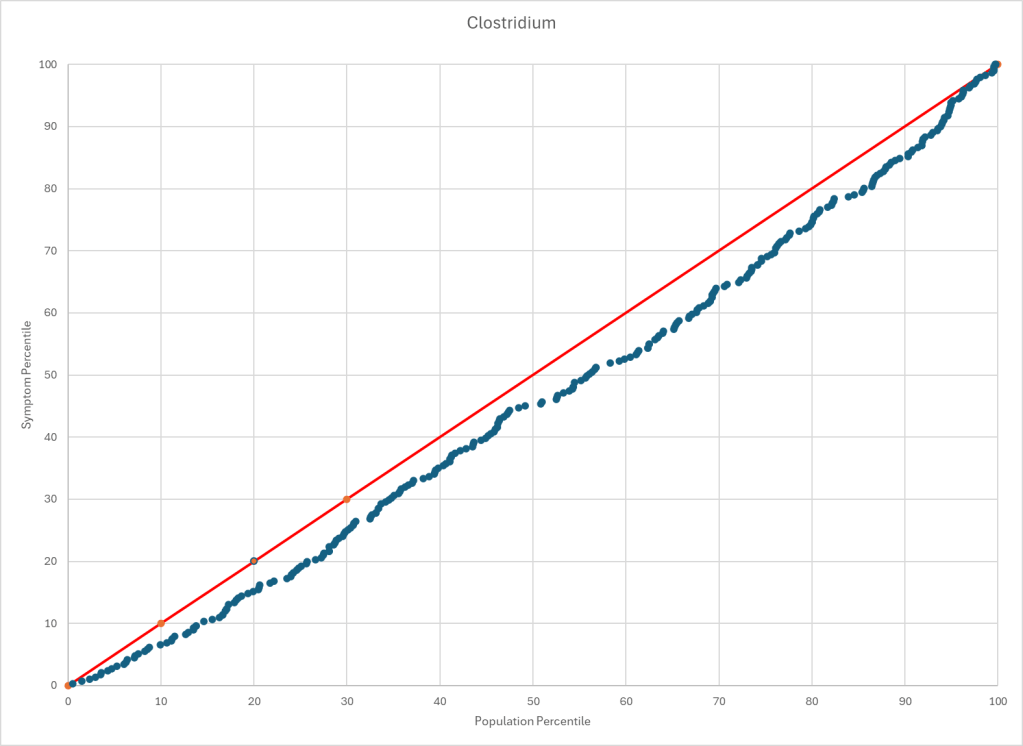
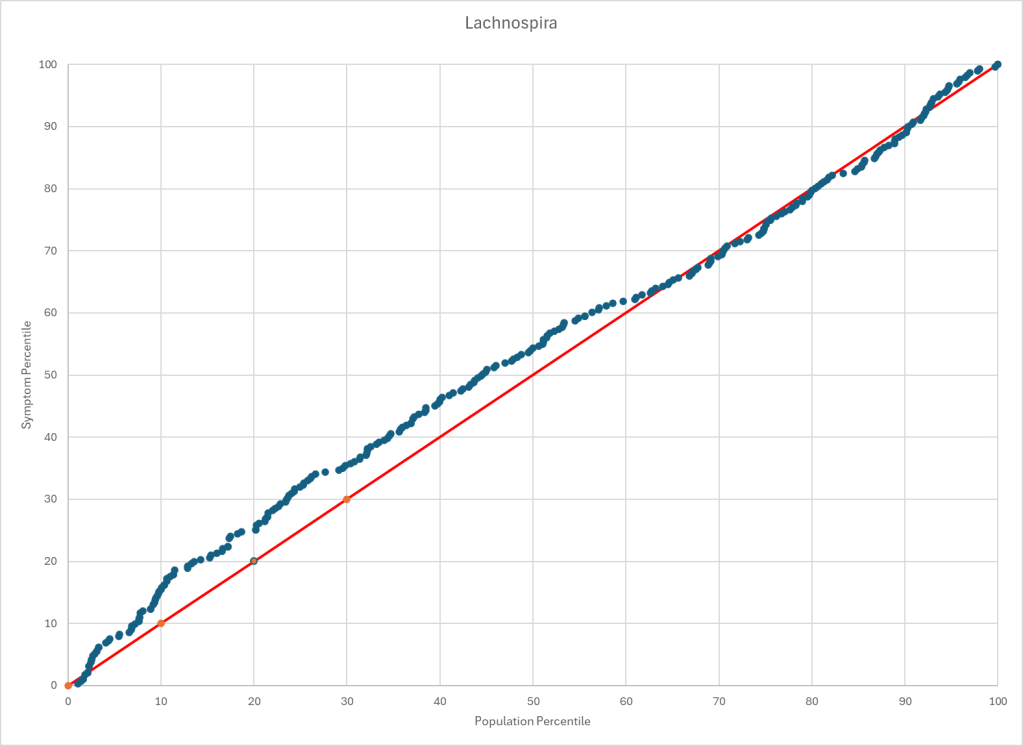
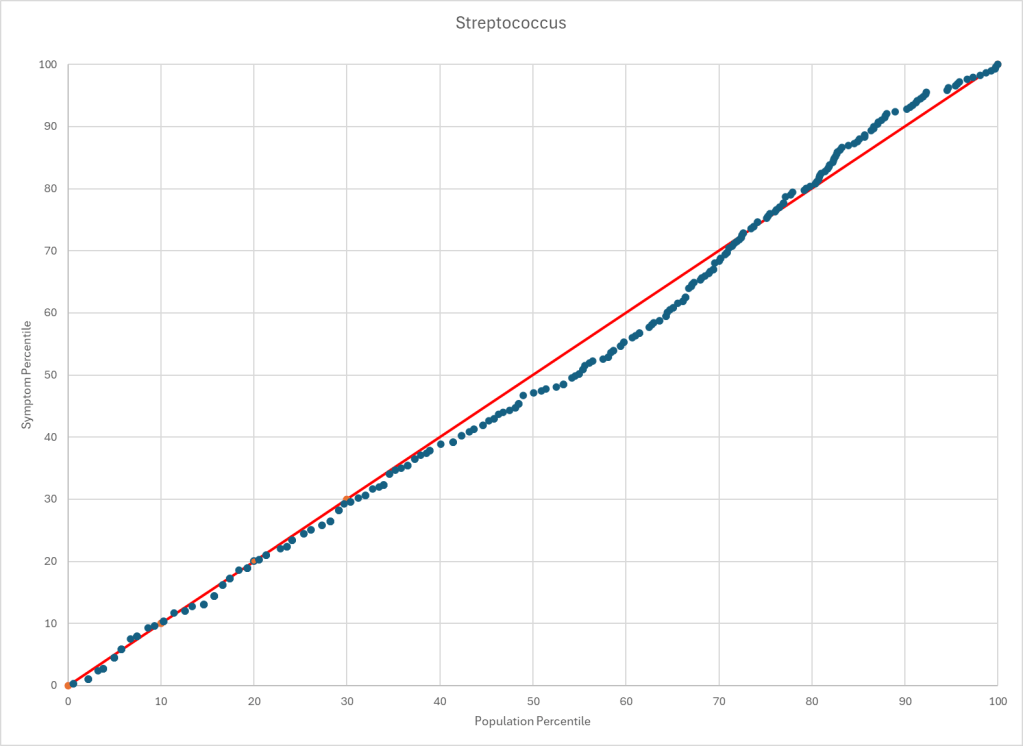
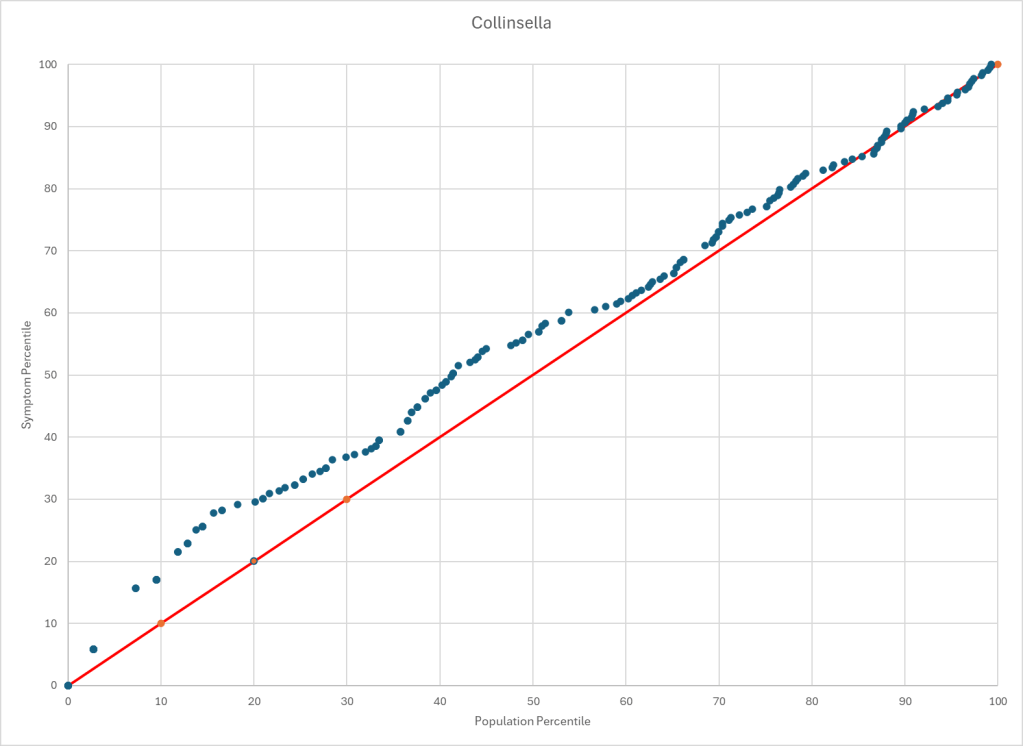
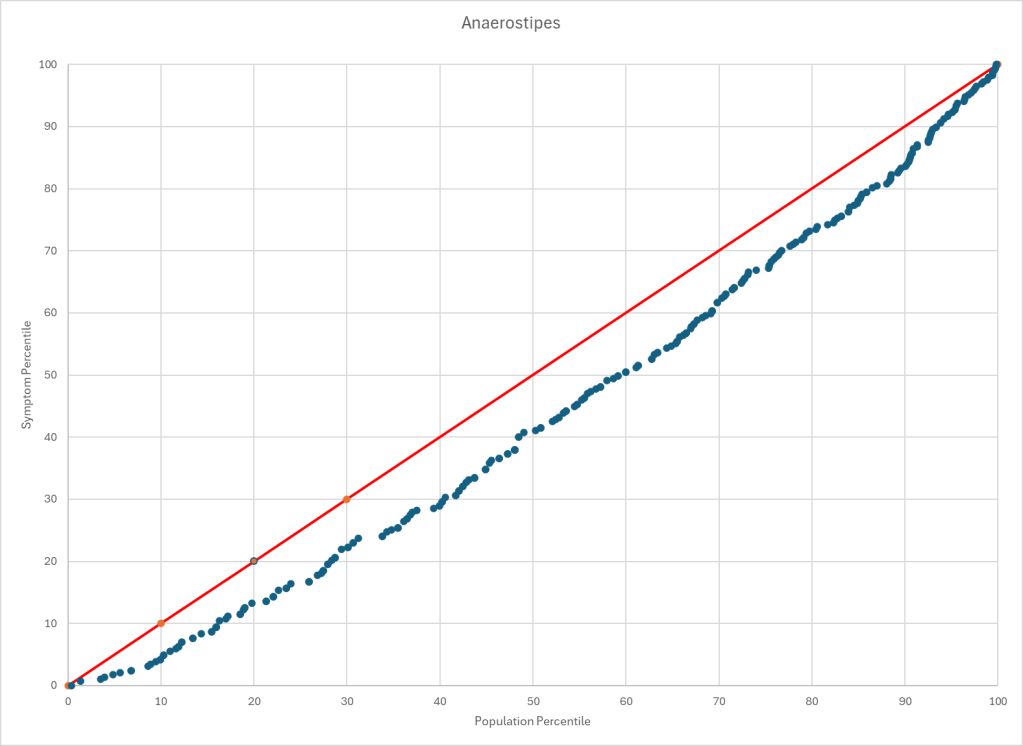
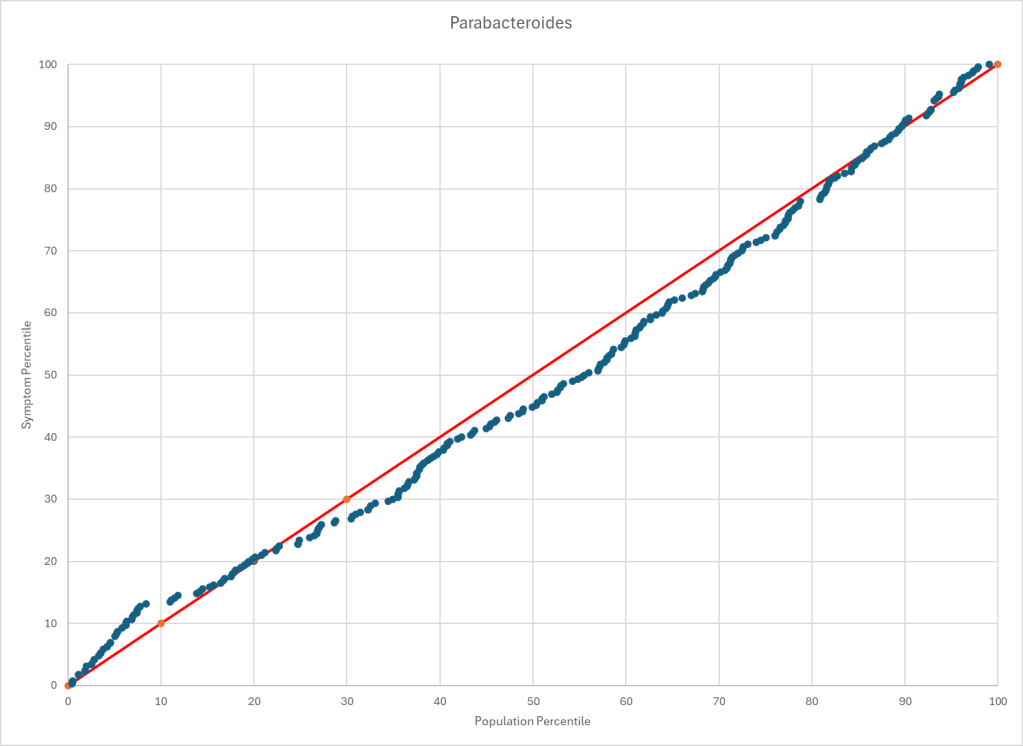
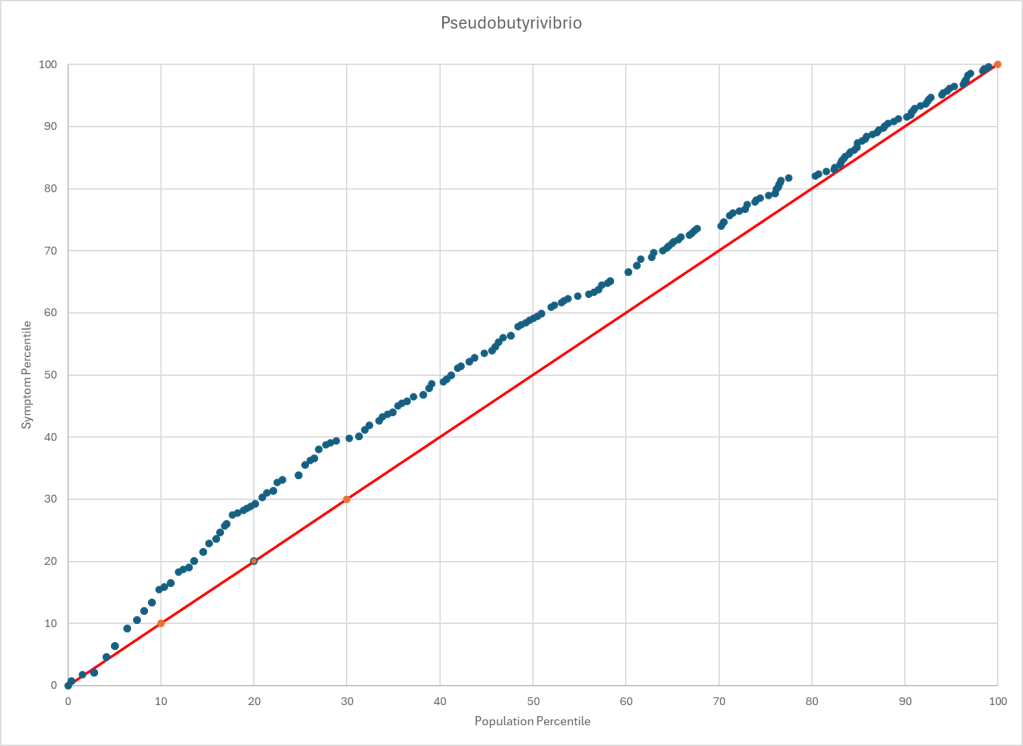
The process is very simple, for a condition like ME/CFS, we compute the expected number of samples reporting this bacteria (based on people without Long COVID) and compare it to the actual number seen. This can be used to compute a statistical value called Chi-Square (χ²), This is then used to compute the chance of it happening at random. This is possible because we have over 3600 samples from some labs and thus able to detect things better.
Actual example:
- Prevotella denticola – Species reported by Ombre
- Expected to see 3.6
- Actually seen 27
- In other words almost 4x more common than expected. The probability is
- 4.9E-34
- or 1 chance in 2,000,000,000,000,000,000,000,000,000,000,000 of happening at random.
- This suggests that we should reduce it to remedy ME/CFS [with the other bacteria involved]
Biomesight and Ombre identifies bacteria using different methodologies so often give different names and amounts. For background on this lack of standardization, see The taxonomy nightmare before Christmas…
The data below is for samples marked with “Official Diagnosis: Chronic Fatigue Syndrome (CFS/ME)“
For Long COVID, see this post.
Unlike some conditions shown below, it is not just one bacteria involved but combinations.
- Peptic ulcer disease: Helicobacter pylori
- Tetanus: Clostridium tetani
- Typhoid fever: Salmonella typhi
- Diphtheria: Corynebacterium diphtheriae
- Syphilis: Treponema pallidum
- Cholera: Vibrio cholerae
- Leprosy: Mycobacterium leprae
- Tuberculosis: Mycobacterium tuberculosis
- Sinusitis: Corynebacterium tuberculostearicum
Ombre Data
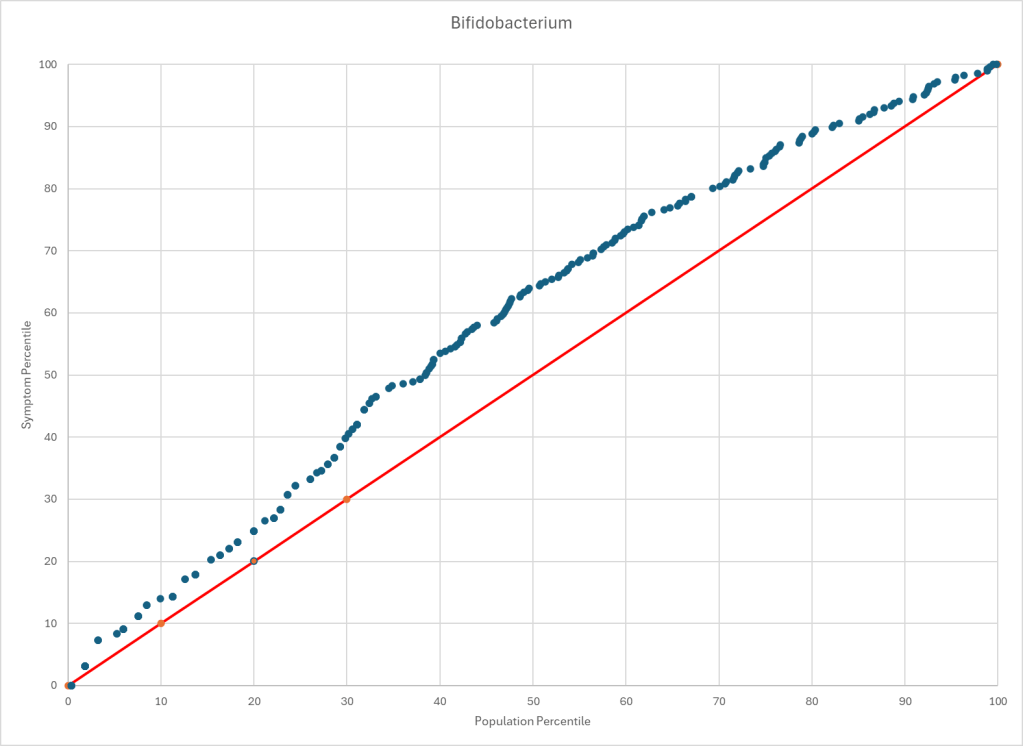
We have 10 bacteria that are too low and some 67 that are too high. Six of the 10 that are too low should be available as probiotics (conceptually), but only B.Bifidum is.
- Bifidobacterium asteroides
- Bifidobacterium bifidum
- Bifidobacterium bombi
- Bifidobacterium commune
- Bifidobacterium magnum
- Bifidobacterium thermacidophilum
In the too high group are some familiar names: Rickettsia (Cecile Jadin’s protocol) and spotted fever group.
| Bacteria Name | Rank | Expected | Observed | Shift | Prob |
| Anaerococcus murdochii | species | 22.1 | 40 | Too High | 0.000134 |
| Anaerococcus octavius | species | 23.2 | 36 | Too High | 0.00785 |
| Anaerococcus senegalensis | species | 21.6 | 34 | Too High | 0.007766 |
| Bacteroides fluxus | species | 82.2 | 111 | Too High | 0.001504 |
| Bacteroides thetaiotaomicron | species | 95.6 | 122 | Too High | 0.006947 |
| Bifidobacterium asteroides | species | 50.3 | 28 | Too Low | 0.004406 |
| Bifidobacterium bifidum | species | 76.3 | 49 | Too Low | 0.002295 |
| Bifidobacterium bombi | species | 80.8 | 44 | Too Low | 0.000136 |
| Bifidobacterium commune | species | 56.8 | 30 | Too Low | 0.000581 |
| Bifidobacterium magnum | species | 73.0 | 46 | Too Low | 0.002755 |
| Bifidobacterium thermacidophilum | species | 53.8 | 31 | Too Low | 0.004352 |
| Campylobacter | genus | 54.0 | 74 | Too High | 0.006445 |
| Campylobacter hominis | species | 26.8 | 43 | Too High | 0.001707 |
| Campylobacteraceae | family | 54.8 | 76 | Too High | 0.004245 |
| Campylobacterales | order | 61.6 | 83 | Too High | 0.006328 |
| Desulfarculaceae | family | 24.9 | 40 | Too High | 0.002442 |
| Desulfarculales | order | 24.9 | 40 | Too High | 0.002442 |
| Desulfarculia | class | 26.2 | 40 | Too High | 0.00692 |
| Desulfocarbo | genus | 14.8 | 26 | Too High | 0.003442 |
| Desulfocarbo indianensis | species | 14.6 | 26 | Too High | 0.002722 |
| Desulfoglaeba | genus | 27.8 | 42 | Too High | 0.007259 |
| Desulfoglaeba alkanexedens | species | 27.4 | 42 | Too High | 0.005339 |
| Desulfovibrio legallii | species | 32.5 | 48 | Too High | 0.006443 |
| Desulfovibrio piger | species | 35.9 | 53 | Too High | 0.004287 |
| Dialister pneumosintes | species | 14.8 | 27 | Too High | 0.001445 |
| Emticicia | genus | 19.3 | 33 | Too High | 0.001758 |
| Emticicia sediminis | species | 8.4 | 21 | Too High | 1.52E-05 |
| Enorma | genus | 22.4 | 35 | Too High | 0.007472 |
| Enorma massiliensis | species | 22.4 | 35 | Too High | 0.007472 |
| Enterorhabdus | genus | 69.6 | 45 | Too Low | 0.004671 |
| Epsilonproteobacteria | class | 62.7 | 84 | Too High | 0.007016 |
| Ezakiella | genus | 44.7 | 63 | Too High | 0.006216 |
| Flavobacteriaceae | family | 75.1 | 98 | Too High | 0.008138 |
| Halanaerobiales | order | 78.4 | 54 | Too Low | 0.005779 |
| Hallella | genus | 34.4 | 60 | Too High | 1.30E-05 |
| Hallella bergensis | species | 17.5 | 39 | Too High | 2.77E-07 |
| Hallella multisaccharivorax | species | 26.2 | 45 | Too High | 0.000235 |
| Holdemania massiliensis | species | 55.2 | 75 | Too High | 0.00788 |
| Hoylesella buccalis | species | 75.1 | 105 | Too High | 0.000552 |
| Hoylesella loescheii | species | 34.3 | 51 | Too High | 0.004268 |
| Hoylesella marshii | species | 15.6 | 30 | Too High | 0.000268 |
| Hoylesella nanceiensis | species | 30.4 | 48 | Too High | 0.001467 |
| Humidesulfovibrio | genus | 29.9 | 49 | Too High | 0.000497 |
| Humidesulfovibrio idahonensis | species | 27.8 | 47 | Too High | 0.000281 |
| Hungateiclostridiaceae | family | 78.0 | 106 | Too High | 0.001492 |
| Insolitispirillum | genus | 31.0 | 46 | Too High | 0.007226 |
| Insolitispirillum peregrinum | species | 30.4 | 46 | Too High | 0.004822 |
| Leptolinea | genus | 14.4 | 26 | Too High | 0.002277 |
| Leptolinea tardivitalis | species | 13.5 | 24 | Too High | 0.004247 |
| Parabacteroides johnsonii | species | 51.0 | 70 | Too High | 0.007925 |
| Paraprevotella xylaniphila | species | 25.0 | 42 | Too High | 0.000677 |
| Parvimonas | genus | 29.7 | 52 | Too High | 4.33E-05 |
| Parvimonas micra | species | 29.7 | 52 | Too High | 4.33E-05 |
| Pedobacter | genus | 36.3 | 53 | Too High | 0.005472 |
| Peptoniphilus lacrimalis | species | 34.6 | 51 | Too High | 0.005246 |
| Persicobacteraceae | family | 12.1 | 24 | Too High | 0.000587 |
| Phocaeicola salanitronis | species | 105.9 | 136 | Too High | 0.003396 |
| Porphyromonas asaccharolytica | species | 33.8 | 51 | Too High | 0.003157 |
| Porphyromonas endodontalis | species | 25.7 | 40 | Too High | 0.004944 |
| Prevotella dentasini | species | 6.8 | 29 | Too High | 1.25E-17 |
| Prevotella denticola | species | 4.2 | 27 | Too High | 1.35E-28 |
| Prolixibacteraceae | family | 20.7 | 33 | Too High | 0.006663 |
| Propioniferax | genus | 23.2 | 42 | Too High | 9.45E-05 |
| Propioniferax innocua | species | 22.8 | 42 | Too High | 5.61E-05 |
| Rickettsia | genus | 21.0 | 36 | Too High | 0.001101 |
| Rickettsia honei | species | 14.8 | 28 | Too High | 0.000569 |
| Segatella bryantii | species | 24.5 | 39 | Too High | 0.003287 |
| Segatella maculosa | species | 33.2 | 55 | Too High | 0.000161 |
| Segatella oris | species | 22.2 | 38 | Too High | 0.000807 |
| Segatella paludivivens | species | 32.9 | 54 | Too High | 0.000245 |
| Senegalimassilia anaerobia | species | 54.0 | 34 | Too Low | 0.006868 |
| Slackia piriformis | species | 56.5 | 29 | Too Low | 0.00048 |
| spotted fever group | species group | 17.9 | 32 | Too High | 0.000907 |
| Tindallia | genus | 14.8 | 26 | Too High | 0.003442 |
| unclassified Burkholderiales | family | 20.9 | 33 | Too High | 0.008034 |
| unclassified Clostridiales | family | 77.1 | 106 | Too High | 0.000984 |
Biomesight Data
We have more data from Biomesight which means better (more) detection of significant bacteria.
As above, we have 12 bacteria that are too low and 116 bacteria that are too high. We have only 2 Bifidobacterium identified with only one available as a probiotic
- Bifidobacterium adolescentis
- Bifidobacterium cuniculi
We see that Lactobacillus is too high. This agrees with brain fog being caused by over production of d-lactic acid.
- Lactobacillus acidophilus
- Lactobacillus iners
We also see these two are high that are commonly associated to issues:
- Streptococcus agalactiae
- Streptococcus mutans
| Tax_Name | Tax_Rank | Expected | Observed | Shift | Probability |
| [Actinobacillus] rossii | species | 119.1 | 89 | Too Low | 0.005858 |
| [Pasteurella] aerogenes-[Pasteurella] mairii-[Actinobacillus] rossii complex | species group | 119.1 | 89 | Too Low | 0.005858 |
| Absiella | genus | 29.8 | 56 | Too High | 1.52E-06 |
| Acetobacteraceae | family | 103.7 | 131 | Too High | 0.007414 |
| Acholeplasma hippikon | species | 73.9 | 110 | Too High | 2.63E-05 |
| Actinobacillus | genus | 176.6 | 134 | Too Low | 0.004701 |
| Actinobacillus pleuropneumoniae | species | 61.6 | 35 | Too Low | 0.00299 |
| Actinobacillus porcinus | species | 133.9 | 99 | Too Low | 0.003018 |
| Adlercreutzia equolifaciens | species | 189.7 | 227 | Too High | 0.006755 |
| Aggregatibacter | genus | 82.3 | 49 | Too Low | 0.000912 |
| Alcanivoracaceae | family | 37.3 | 54 | Too High | 0.006427 |
| Alcanivorax | genus | 37.3 | 54 | Too High | 0.006427 |
| Amedibacillus | genus | 196.8 | 235 | Too High | 0.006495 |
| Amedibacillus dolichus | species | 197.1 | 235 | Too High | 0.006912 |
| Anaerococcus | genus | 126.4 | 157 | Too High | 0.006444 |
| Anaerococcus hydrogenalis | species | 39.1 | 58 | Too High | 0.002475 |
| Anaerococcus prevotii | species | 21.1 | 36 | Too High | 0.001188 |
| Anaerococcus tetradius | species | 29.6 | 59 | Too High | 6.37E-08 |
| Anaerococcus vaginalis | species | 63.6 | 87 | Too High | 0.003269 |
| Anaerofustis | genus | 54.3 | 76 | Too High | 0.003316 |
| Anaerofustis stercorihominis | species | 53.0 | 74 | Too High | 0.003981 |
| Arthrobacter | genus | 27.7 | 43 | Too High | 0.003655 |
| Bifidobacterium adolescentis | strain | 102.4 | 63 | Too Low | 0.001744 |
| Bifidobacterium cuniculi | species | 64.9 | 38 | Too Low | 0.007829 |
| Bifidobacterium ruminantium | species | 14.8 | 30 | Too High | 7.46E-05 |
| Caloramator viterbiensis | species | 19.0 | 37 | Too High | 3.60E-05 |
| Campylobacter ureolyticus | species | 38.0 | 56 | Too High | 0.003481 |
| Cetobacterium | genus | 17.7 | 29 | Too High | 0.007331 |
| Cetobacterium ceti | species | 13.7 | 28 | Too High | 0.000115 |
| Chlorobaculum limnaeum | species | 8.6 | 22 | Too High | 4.66E-06 |
| Clostridium cadaveris | species | 106.5 | 140 | Too High | 0.001155 |
| Collinsella tanakaei | species | 43.8 | 62 | Too High | 0.005945 |
| Corynebacteriaceae | family | 133.0 | 164 | Too High | 0.007125 |
| Corynebacterium | genus | 133.0 | 164 | Too High | 0.007125 |
| Corynebacterium amycolatum | species | 20.6 | 36 | Too High | 0.000675 |
| Corynebacterium glucuronolyticum | species | 10.4 | 21 | Too High | 0.001029 |
| Corynebacterium pyruviciproducens | species | 12.7 | 23 | Too High | 0.003812 |
| Corynebacterium xerosis | species | 14.8 | 25 | Too High | 0.007807 |
| Dehalobacterium | genus | 124.4 | 159 | Too High | 0.001898 |
| Dehalococcoidaceae | family | 11.9 | 23 | Too High | 0.00124 |
| Dehalococcoidales | order | 11.9 | 23 | Too High | 0.00124 |
| Dehalococcoidia | class | 11.9 | 23 | Too High | 0.00124 |
| Dehalogenimonas | genus | 11.9 | 23 | Too High | 0.00124 |
| Dehalogenimonas lykanthroporepellens | species | 11.3 | 23 | Too High | 0.000539 |
| Desulfonatronovibrio | genus | 44.2 | 62 | Too High | 0.007398 |
| Desulfonatronovibrionaceae | family | 43.3 | 62 | Too High | 0.004437 |
| Desulfovibrio psychrotolerans | species | 19.8 | 37 | Too High | 0.000109 |
| Dethiobacteraceae | family | 47.8 | 77 | Too High | 2.50E-05 |
| Dethiobacterales | order | 47.8 | 77 | Too High | 2.50E-05 |
| Dethiobacteria | class | 47.8 | 77 | Too High | 2.50E-05 |
| Dyadobacter | genus | 77.6 | 103 | Too High | 0.003878 |
| Eggerthella sinensis | species | 133.2 | 167 | Too High | 0.003441 |
| Erysipelothrix inopinata | species | 42.5 | 62 | Too High | 0.002739 |
| Ethanoligenens | genus | 158.8 | 205 | Too High | 0.000248 |
| Eubacterium limosum | species | 27.2 | 42 | Too High | 0.00458 |
| Facklamia | genus | 21.2 | 34 | Too High | 0.005361 |
| Filifactor alocis | species | 35.1 | 54 | Too High | 0.001384 |
| Finegoldia magna | species | 112.9 | 143 | Too High | 0.004643 |
| Halanaerobiaceae | family | 56.2 | 78 | Too High | 0.003557 |
| Halanaerobiales | order | 61.2 | 83 | Too High | 0.005272 |
| Halanaerobium | genus | 56.2 | 78 | Too High | 0.003557 |
| Haploplasma | genus | 13.9 | 31 | Too High | 4.32E-06 |
| Haploplasma cavigenitalium | species | 13.9 | 31 | Too High | 4.32E-06 |
| Heliorestis baculata | species | 17.4 | 30 | Too High | 0.002634 |
| Hungateiclostridiaceae | family | 45.4 | 71 | Too High | 0.000143 |
| Hungateiclostridium | genus | 45.2 | 71 | Too High | 0.000124 |
| Hyphomicrobium | genus | 39.8 | 58 | Too High | 0.004009 |
| Hyphomicrobium aestuarii | species | 34.0 | 51 | Too High | 0.003635 |
| Hyphomonadaceae | family | 16.9 | 29 | Too High | 0.003196 |
| Hyphomonadales | order | 16.9 | 29 | Too High | 0.003196 |
| Isoalcanivorax | genus | 35.5 | 52 | Too High | 0.005689 |
| Isoalcanivorax indicus | species | 31.1 | 52 | Too High | 0.000184 |
| Lactobacillus acidophilus | species | 18.8 | 32 | Too High | 0.002356 |
| Lactobacillus iners | species | 17.3 | 32 | Too High | 0.000436 |
| Legionella shakespearei | species | 67.0 | 91 | Too High | 0.003402 |
| Levilactobacillus | genus | 12.2 | 25 | Too High | 0.000263 |
| Listeriaceae | family | 178.9 | 215 | Too High | 0.006915 |
| Luteolibacter | genus | 127.8 | 159 | Too High | 0.005844 |
| Luteolibacter algae | species | 126.9 | 158 | Too High | 0.005805 |
| Mannheimia | genus | 134.7 | 102 | Too Low | 0.004859 |
| Mannheimia caviae | species | 128.7 | 97 | Too Low | 0.005142 |
| Meiothermus | genus | 78.6 | 103 | Too High | 0.005964 |
| Meiothermus granaticius | species | 77.3 | 101 | Too High | 0.007138 |
| Mobiluncus | genus | 31.7 | 52 | Too High | 0.0003 |
| Mogibacterium vescum | species | 57.6 | 94 | Too High | 1.64E-06 |
| Nannocystineae | suborder | 23.0 | 36 | Too High | 0.006465 |
| Olivibacter terrae | species | 12.1 | 22 | Too High | 0.004168 |
| Paenibacillus filicis | species | 22.9 | 38 | Too High | 0.001632 |
| Pasteurellaceae incertae sedis | no rank | 119.1 | 89 | Too Low | 0.005858 |
| Pediococcus | genus | 166.2 | 208 | Too High | 0.00119 |
| Pediococcus argentinicus | species | 17.9 | 31 | Too High | 0.002047 |
| Peptoniphilus asaccharolyticus | species | 116.8 | 148 | Too High | 0.003873 |
| Peptostreptococcus stomatis | species | 52.0 | 73 | Too High | 0.003541 |
| Porphyromonas asaccharolytica | species | 70.2 | 101 | Too High | 0.000234 |
| Prosthecobacter | genus | 32.7 | 54 | Too High | 0.000194 |
| Prosthecobacter fluviatilis | species | 18.1 | 33 | Too High | 0.00045 |
| Rhizobiaceae | family | 80.9 | 105 | Too High | 0.007378 |
| Roseomonas | genus | 22.3 | 38 | Too High | 0.000867 |
| Roseospira | genus | 54.9 | 78 | Too High | 0.0018 |
| Ruminiclostridium cellulolyticum | species | 107.1 | 135 | Too High | 0.007053 |
| Segatella paludivivens | species | 72.9 | 45 | Too Low | 0.001097 |
| Slackia faecicanis | species | 98.6 | 128 | Too High | 0.003086 |
| Sporosarcina | genus | 47.9 | 69 | Too High | 0.00235 |
| Sporosarcina pasteurii | species | 46.9 | 69 | Too High | 0.001278 |
| Streptococcus agalactiae | species | 11.9 | 23 | Too High | 0.00124 |
| Streptococcus mutans | species | 31.7 | 51 | Too High | 0.0006 |
| Thauera terpenica | species | 20.6 | 33 | Too High | 0.006179 |
| Thermaceae | family | 94.7 | 125 | Too High | 0.001859 |
| Thermales | order | 94.7 | 125 | Too High | 0.001859 |
| Thermoanaerobacter | genus | 78.4 | 102 | Too High | 0.007565 |
| Thermodesulfatator | genus | 22.7 | 42 | Too High | 5.03E-05 |
| Thermodesulfatator atlanticus | species | 22.7 | 42 | Too High | 5.03E-05 |
| Thermodesulfatatoraceae | family | 27.5 | 42 | Too High | 0.005627 |
| Thermodesulfobacteria | class | 22.7 | 42 | Too High | 5.03E-05 |
| Thermodesulfobacteriaceae | family | 22.7 | 42 | Too High | 5.03E-05 |
| Thermodesulfobacteriales | order | 22.7 | 42 | Too High | 5.03E-05 |
| Thermosediminibacteraceae | family | 14.2 | 26 | Too High | 0.001847 |
| Thermovenabulum | genus | 14.2 | 26 | Too High | 0.001847 |
| Thermovenabulum ferriorganovorum | species | 14.2 | 26 | Too High | 0.001847 |
| Thermus | genus | 28.5 | 43 | Too High | 0.006576 |
| Thiocapsa | genus | 75.7 | 104 | Too High | 0.001154 |
| unclassified Burkholderiales | family | 15.2 | 29 | Too High | 0.000429 |
| Vagococcus teuberi | species | 47.0 | 66 | Too High | 0.005467 |
| Varibaculum | genus | 47.5 | 73 | Too High | 0.000214 |
| Varibaculum cambriense | species | 19.3 | 32 | Too High | 0.003695 |
| Winkia | genus | 11.0 | 23 | Too High | 0.000324 |
| Winkia neuii | species | 11.0 | 23 | Too High | 0.000324 |
uBiome
While the company no longer exists, we have a significant number of samples from them.
As above, we have Lactobacillus crispatus being too high. We have just 2 bacteria being too low and 96 being too high.
| Tax_Name | Tax_Rank | Expected | Observed | Shift | Probability |
| Acetitomaculum | genus | 24.4 | 44 | Too High | 7.07E-05 |
| Acholeplasmatales | order | 10.0 | 21 | Too High | 0.000503 |
| Actinomyces sp. 2002-2301122 | species | 14.4 | 25 | Too High | 0.00497 |
| Aggregatibacter | genus | 42.2 | 24 | Too Low | 0.005171 |
| Alistipes indistinctus | species | 62.8 | 90 | Too High | 0.000602 |
| Alistipes sp. NML05A004 | species | 81.8 | 111 | Too High | 0.001227 |
| Alistipes sp. RMA 9912 | species | 13.7 | 25 | Too High | 0.002304 |
| Anaerococcus murdochii | species | 30.5 | 56 | Too High | 3.93E-06 |
| Anaeroplasmataceae | family | 22.8 | 38 | Too High | 0.00148 |
| Anaeroplasmatales | order | 23.1 | 39 | Too High | 0.000914 |
| Archaea | superkingdom | 39.7 | 73 | Too High | 1.32E-07 |
| Atopobium | genus | 17.8 | 30 | Too High | 0.00374 |
| Bacteroides eggerthii | species | 30.5 | 52 | Too High | 9.98E-05 |
| Bacteroides sp. 35AE37 | species | 66.0 | 92 | Too High | 0.00139 |
| Bacteroides sp. XB12B | species | 39.7 | 66 | Too High | 3.10E-05 |
| Blautia stercoris | species | 37.2 | 55 | Too High | 0.003458 |
| Butyricimonas | genus | 88.4 | 114 | Too High | 0.006407 |
| Butyricimonas faecihominis | species | 57.7 | 79 | Too High | 0.005005 |
| Butyricimonas sp. 214-4 | species | 53.8 | 75 | Too High | 0.003921 |
| Butyricimonas virosa | species | 42.3 | 62 | Too High | 0.002455 |
| Caldicoprobacter | genus | 23.9 | 39 | Too High | 0.001957 |
| Caldicoprobacteraceae | family | 44.2 | 68 | Too High | 0.00034 |
| Campylobacter hominis | species | 27.9 | 42 | Too High | 0.007781 |
| Campylobacter ureolyticus | species | 24.4 | 41 | Too High | 0.000762 |
| Catenibacterium | genus | 57.9 | 80 | Too High | 0.003678 |
| Christensenella minuta | species | 23.1 | 36 | Too High | 0.007121 |
| Cloacibacillus | genus | 19.8 | 38 | Too High | 4.36E-05 |
| Cloacibacillus evryensis | species | 14.7 | 31 | Too High | 2.24E-05 |
| Coprobacter secundus | species | 32.8 | 53 | Too High | 0.000426 |
| Corynebacterium diphtheriae | species | 11.5 | 21 | Too High | 0.005334 |
| Corynebacterium sp. 713182/2012 | species | 16.2 | 27 | Too High | 0.006945 |
| Desulfovibrio | genus | 72.6 | 97 | Too High | 0.004103 |
| Desulfovibrio sp. 3_1_syn3 | species | 9.0 | 30 | Too High | 2.23E-12 |
| Dialister micraerophilus | species | 9.6 | 24 | Too High | 3.85E-06 |
| Dialister propionicifaciens | species | 41.0 | 58 | Too High | 0.008017 |
| Dialister sp. S7MSR5 | species | 16.8 | 29 | Too High | 0.002792 |
| Eubacterium | genus | 27.2 | 46 | Too High | 0.000305 |
| Euryarchaeota | phylum | 39.7 | 73 | Too High | 1.32E-07 |
| Fibrobacter | genus | 14.9 | 27 | Too High | 0.001656 |
| Fibrobacteraceae | family | 24.1 | 47 | Too High | 3.08E-06 |
| Fibrobacterales | order | 32.6 | 61 | Too High | 6.22E-07 |
| Fibrobacteria | class | 32.6 | 61 | Too High | 6.22E-07 |
| Fibrobacterota | phylum | 32.6 | 61 | Too High | 6.22E-07 |
| Gelria | genus | 37.6 | 65 | Too High | 7.75E-06 |
| Granulicatella adiacens | species | 69.5 | 45 | Too Low | 0.006077 |
| Herbaspirillum | genus | 46.7 | 72 | Too High | 0.000218 |
| Herbaspirillum seropedicae | species | 39.0 | 62 | Too High | 0.000225 |
| Hespellia | genus | 96.4 | 123 | Too High | 0.006732 |
| Hoylesella timonensis | species | 23.4 | 40 | Too High | 0.000577 |
| Hungateiclostridiaceae | family | 26.4 | 43 | Too High | 0.001246 |
| Hydrogenoanaerobacterium | genus | 40.6 | 64 | Too High | 0.000246 |
| Lactobacillus crispatus | species | 26.1 | 40 | Too High | 0.006759 |
| Lentisphaeria | class | 53.6 | 75 | Too High | 0.003432 |
| Lentisphaerota | phylum | 53.6 | 75 | Too High | 0.003432 |
| Methanobacteria | class | 37.4 | 73 | Too High | 6.10E-09 |
| Methanobacteriaceae | family | 37.4 | 73 | Too High | 6.10E-09 |
| Methanobacteriales | order | 37.4 | 73 | Too High | 6.10E-09 |
| Methanobrevibacter | genus | 36.9 | 71 | Too High | 2.03E-08 |
| Methanobrevibacter smithii | species | 36.4 | 66 | Too High | 9.34E-07 |
| Methanomada group | clade | 37.4 | 73 | Too High | 6.10E-09 |
| Mollicutes | class | 30.0 | 49 | Too High | 0.00052 |
| Murdochiella | genus | 42.2 | 65 | Too High | 0.000434 |
| Mycoplasmatota | phylum | 30.0 | 49 | Too High | 0.00052 |
| Opitutia | class | 16.4 | 33 | Too High | 4.20E-05 |
| Oxalobacteraceae | family | 47.7 | 73 | Too High | 0.000257 |
| Parabacteroides johnsonii | species | 18.8 | 34 | Too High | 0.000451 |
| Parasporobacterium paucivorans | species | 16.2 | 29 | Too High | 0.001388 |
| Parvibacter | genus | 14.1 | 33 | Too High | 4.82E-07 |
| Parvibacter caecicola | species | 12.3 | 29 | Too High | 1.95E-06 |
| Peptococcus | genus | 71.8 | 100 | Too High | 0.000867 |
| Peptoniphilus lacrimalis | species | 30.3 | 46 | Too High | 0.004193 |
| Porphyromonas sp. 2024b | species | 12.7 | 29 | Too High | 4.76E-06 |
| Propionibacteriaceae | family | 18.8 | 31 | Too High | 0.004859 |
| Propionibacteriales | order | 18.8 | 31 | Too High | 0.004859 |
| Puniceicoccales | order | 16.2 | 33 | Too High | 2.76E-05 |
| Robinsoniella | genus | 30.3 | 50 | Too High | 0.00033 |
| Robinsoniella sp. KNHs210 | species | 22.6 | 37 | Too High | 0.002366 |
| Staphylococcaceae | family | 38.5 | 57 | Too High | 0.002786 |
| Staphylococcus | genus | 38.2 | 57 | Too High | 0.00235 |
| Synergistaceae | family | 41.8 | 72 | Too High | 2.96E-06 |
| Synergistales | order | 41.8 | 72 | Too High | 2.96E-06 |
| Synergistia | class | 41.8 | 72 | Too High | 2.96E-06 |
| Synergistota | phylum | 41.8 | 72 | Too High | 2.96E-06 |
| Thermoanaerobacteraceae | family | 37.7 | 65 | Too High | 8.62E-06 |
| Thermoanaerobacterales | order | 39.7 | 66 | Too High | 3.10E-05 |
| unclassified Butyricimonas | no rank | 53.1 | 77 | Too High | 0.001019 |
| unclassified Desulfovibrio | no rank | 12.6 | 43 | Too High | 8.86E-18 |
| unclassified Finegoldia | no rank | 31.8 | 47 | Too High | 0.006982 |
| unclassified Phascolarctobacterium | no rank | 11.5 | 23 | Too High | 0.000738 |
| unclassified Porphyromonas | no rank | 24.4 | 38 | Too High | 0.005801 |
| unclassified Prevotella | no rank | 9.6 | 21 | Too High | 0.000258 |
| unclassified Robinsoniella | no rank | 22.6 | 37 | Too High | 0.002366 |
| unclassified Staphylococcus | no rank | 26.4 | 43 | Too High | 0.001246 |
| Varibaculum | genus | 48.5 | 70 | Too High | 0.001966 |
| Varibaculum sp. CCUG 45114 | species | 36.1 | 54 | Too High | 0.002986 |
| Victivallaceae | family | 53.6 | 75 | Too High | 0.003432 |
| Victivallales | order | 53.6 | 75 | Too High | 0.003432 |
| Victivallis | genus | 53.1 | 75 | Too High | 0.002607 |
Bottom Line
In terms of probiotics, there are two that should be considered:
- Bif. Bifidum
- Bif. Adolescentis
And all Lactobacillus probiotics avoided.
The above information will be eventually integrated into Microbiome Prescription suggestions expert system. The purpose is to first identify the bacteria of concern.
The following bacteria were reported by 2 or 3 of the above
| Aggregatibacter | genus |
| Anaerococcus murdochii | species |
| Campylobacter hominis | species |
| Campylobacter ureolyticus | species |
| Halanaerobiales | order |
| Hungateiclostridiaceae | family |
| Parabacteroides johnsonii | species |
| Peptoniphilus lacrimalis | species |
| Porphyromonas asaccharolytica | species |
| Segatella paludivivens | species |
| unclassified Burkholderiales | family |
| Varibaculum | genus |