I have been observing the Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) community for almost 35 years. I have been an active member of this community. I have also been in remission for it, but deeply interested in understanding it; I have had several relapses. I wanted to prevent future relapses hence great interest in reading and researching.
Microbiome Analysis as a path with good results
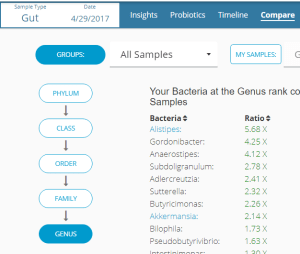
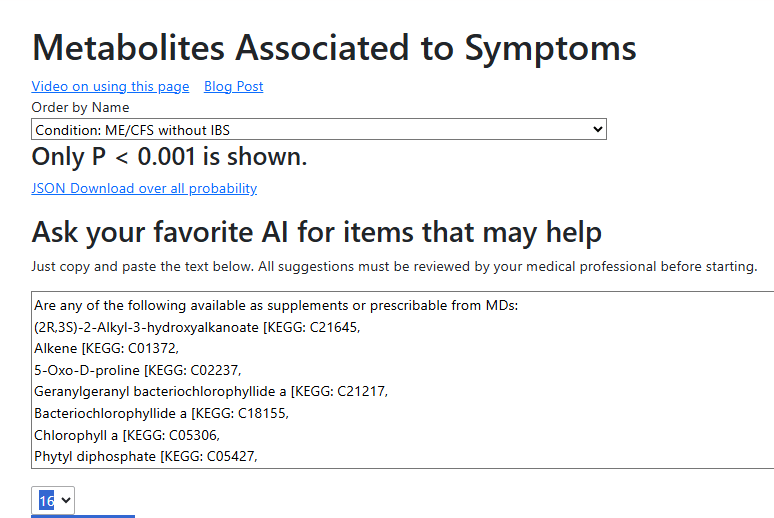
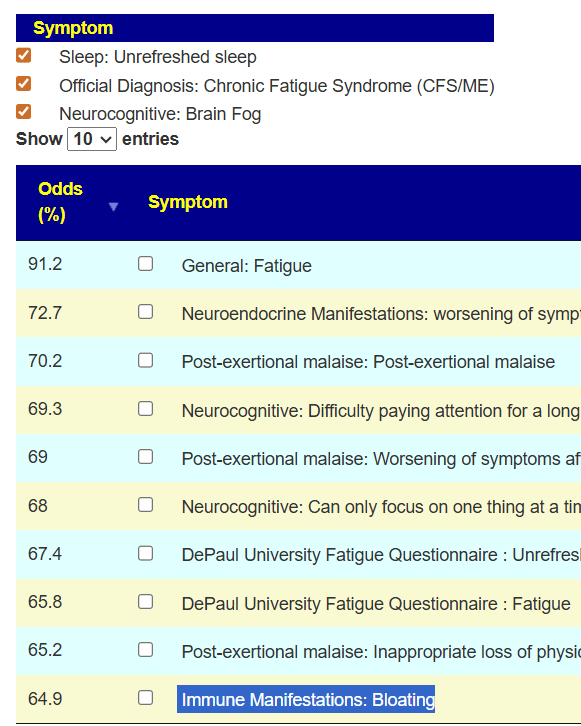
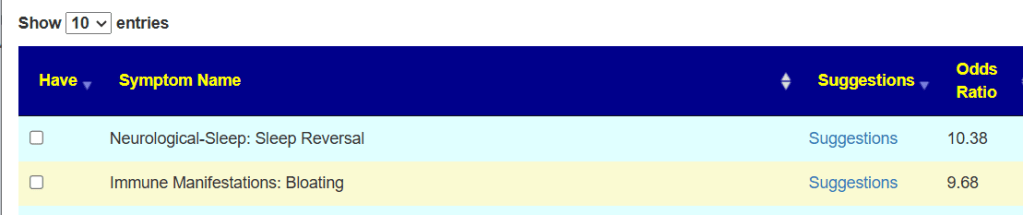
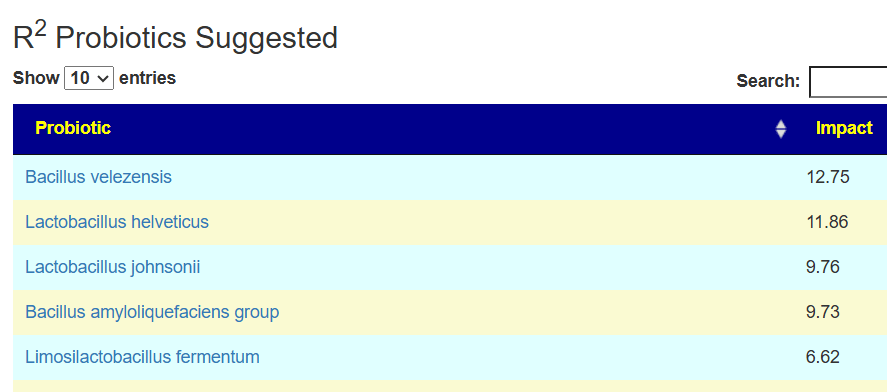
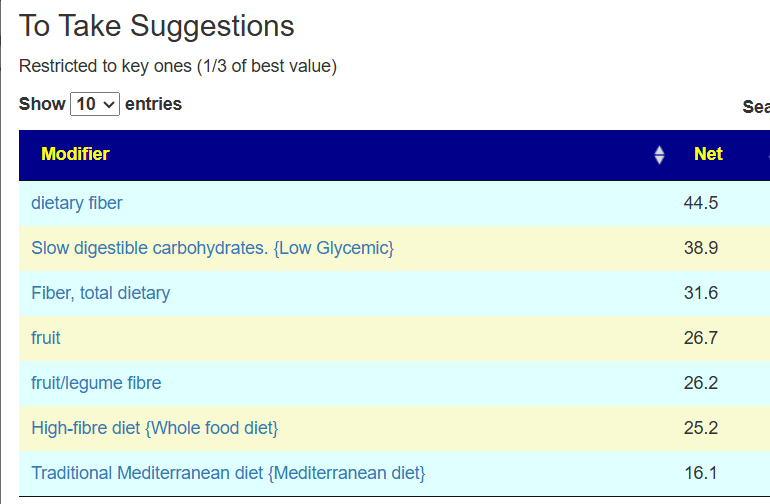
My main focus in 2023 is on the microbiome of ME/CFS people. I have a collections of reviews of individual microbiome samples reviews available. The data from the suggested tests are processed using the free expert system at Microbiome Prescription. So far, every person who has done this route and repeated the microbiome tests have reported subject improvement and objective improvement. It can be a long journey, we take one step at a time.
During those decades I have seen a huge number of theories, suggested treatments and clinical findings. I have seen many “this cured me” which failed to deliver remission for anyone else. I have seen treatment plans proposed / sold by well-meaning MDs that worsen the condition and persisted for years. I see this still going on today for both ME/CFS and Long Hauler COVID.
My training is in science: mathematics, statistics, general sciences. I have been an instructor in those areas at many universities, colleges and even high schools. My remissions resulted from following gold or silver standard evidence. I have read several thousand published papers on ME/CFS (with and without brain fog).
What is on this site?
Most of the posts results from reading a new paper, readers asking questions, seeing a question asked on some online groups. The posts follow this pattern:
- The question or concern that started the post
- A review of all literature on the US National Library of Medicine, conference papers, etc.
- Quotes provided from papers or their titles
- Links to the paper so people or their medical professionals can read for themselves
- A bottom line section which are my conclusions inferred from the literature.
To quote Seargent Friday from the old Dragnet TV Series, “All we want are the facts, ma’am“.
Use the site as an Encyclopedia
There is another similar site, Encyclopedia Myalgic encephalomyelitis, [me-pedia.org], which I also use. I actually have had more page views than that site! What the difference? I seem to cover things in more depth. I just searched for “grapeseed” on that site and found nothing. I searched on this site and literally had dozens of posts cited. Resveratrol Revisited [2015], Resveratrol Recap [2017], Mast Cell Moderators — non-drugs and suspect bacteria [2023]
The fastest way to find information is to go to google search, enter site:cfsremission.com followed by what you are interested in.
I Promote No Protocol, I promote evidence based treatment.
As a statistician, I recognized that ME/CFS consists of many, many, many, different subsets. To treat successfully means getting information. I know that my own ME/CFS was usually triggered by the interaction of an inherited coagulation defect (Prothrombin G20210A a.k.a. Factor II Mutation) interacting with stress and a microbiome that goes bad with stress. What works for me may not work for another with ME/CFS.
My Unified Model of ME/CFS and Long COVID
I have a hypothesis on the causes of ME/CFS and a treatment approach (not a protocol) that is likely to reduce the severity of ME/CFS. Some may go into remission.
The Cause of ME/CFS
The cause is rather simple: anything that causes an alteration of the microbiome. This may be food poisoning , an infection (COVID, Flu, Lyme, Epstein-Barr virus), a vaccination, a prescription or over the counter drug, pesticides, bad diet. This alteration and happenstance cascade into a microbiome dysfunction that mucks up your system. Too much of some metabolites /chemicals are being produced, too little of others, the chemicals used by the body are “hacked” and the body manifests a huge variety of symptoms.
Treatment Approach
Over the 30+ years, I have been a good mathematical modeler. Trying to find a model that agrees with all of the facts, explains all of the observations and last, can make predictions that are testable. Not that many years ago I came to the realization that the microbiome dysregulation model was an extremely good fit. More important, it made predictions that could be tested.
A 1999 study in Australia found a common pattern with ME/CFS patients microbiome. If something helps ME/CFS then it is likely that this same thing would influence the bacteria found to move closer to normal. This post from 2013, Finally! Someone verified the 1999 Studies – Thank you Kenny De Meirleir! shifted me further down this path. Finding that the most effective antibiotic protocols for ME/CFS and Lyme would also correct the reported microbiome function (see Why Jadin’s Antibiotics Protocol usually work — Pasteur Institute got the solution right and the explanation wrong? ).
I keep monitoring ME/CFS studies. My current effort is dealing with improving the ability to correct the microbiome. This is done on a separate site, Microbiome Prescription, and a separate blog site.
I have a page linking to people experience with this approach. Each person is different.
Analysis Posts on Long COVID and ME/CFS
I believe that most ME/CFS people will improve significantly with microbiome testing followed by appropriate adjustments. This is often an iterative approach (test-adjust-repeat).